Throughout replication, expression, and restore of the eukaryotic genome, mobile equipment should entry the DNA wrapped round histone proteins forming nucleosomes. These octameric protein·DNA complexes are modular, dynamic, and versatile and unwrap or disassemble both spontaneously or by the motion of molecular motors. Thus, the mechanism of formation and regulation of subnucleosomal intermediates has gained consideration genome-wide as a result of it controls DNA accessibility.*
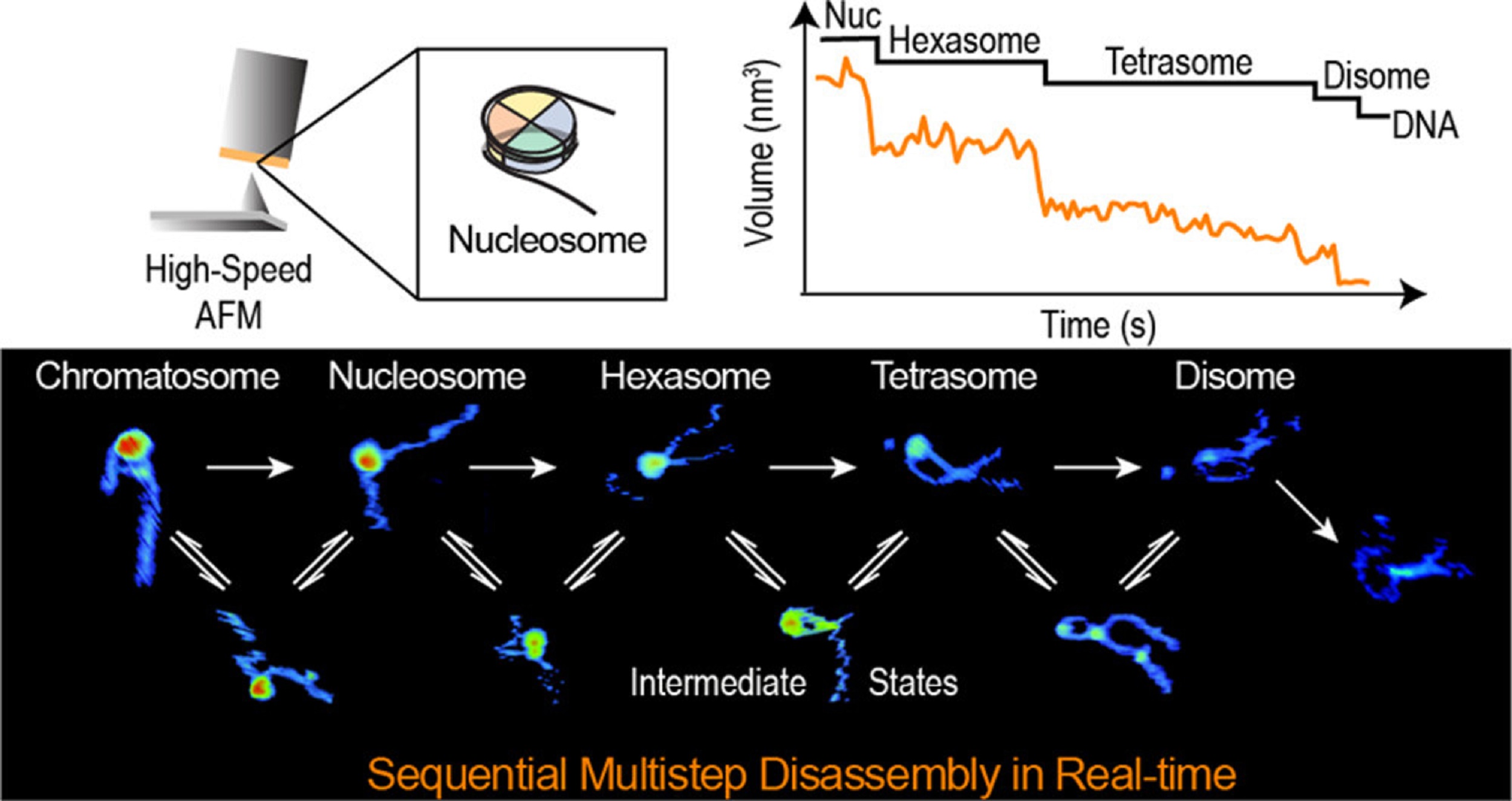
Within the article “Actual-Time Multistep Asymmetrical Disassembly of Nucleosomes and Chromatosomes Visualized by Excessive-Pace Atomic Power Microscopy” Bibiana Onoa, César Díaz-Celis, Cristhian Cañari-Chumpitaz, Antony Lee and Carlos Bustamante describe how they imaged nucleosomes and their extra compacted construction with the linker histone H1 (chromatosomes) utilizing high-speed atomic pressure microscopy to visualise concurrently the modifications within the DNA and the histone core throughout their disassembly when deposited on mica.*
Moreover, Bibiana Onoa et al. skilled a neural community and developed an computerized algorithm to trace molecular structural modifications in actual time. *
The authors’ outcomes present that nucleosome disassembly is a sequential course of involving asymmetrical stepwise dimer ejection occasions. The presence of H1 restricts DNA unwrapping, considerably will increase the nucleosomal lifetime, and impacts the pathway by which heterodimer asymmetrical dissociation happens. *
Bibiana Onoa et al. observe that tetrasomes are resilient to disassembly and that the tetramer core (H3·H4)2 can diffuse alongside the nucleosome positioning sequence. Tetrasome mobility is likely to be essential to the correct meeting of nucleosomes and will be related throughout nucleosomal transcription, as tetrasomes survive RNA polymerase passage. These findings are related to understanding nucleosome intrinsic dynamics and their modification by DNA-processing enzymes. *
To characterize the nucleosomes dynamics in 2D, particular person molecules have been noticed in buffer utilizing an Ando-type excessive pace atomic pressure microscope along with NanoWorld Extremely-Brief Cantilevers for HS-AFM of the USC-F1.2-K0.15 AFM probe kind ( typical spring fixed 0.15 N/m, typical resonance frequency in air 1200 kHz, resonance frequency 500–600 kHz in liquid). *
The AFM information offered within the article permit the authors to instantly visualize the dynamics of DNA and histones throughout nucleosome and chromatosome disassembly, offering a simultaneous statement of DNA unwrapping and histone dissociation. *
The experimental and analytical technique offered exhibits that real-time HS-AFM is a sturdy and highly effective software for finding out single nucleosomes and chromatin dynamics. *
*Bibiana Onoa, César Díaz-Celis, Cristhian Cañari-Chumpitaz, Antony Lee and Carlos Bustamante
Actual-Time Multistep Asymmetrical Disassembly of Nucleosomes and Chromatosomes Visualized by Excessive-Pace Atomic Power Microscopy
ACS Central Science 2024, 10, 1, 122–137
DOI: https://doi.org/10.1021/acscentsci.3c00735
Open Entry The article “Actual-Time Multistep Asymmetrical Disassembly of Nucleosomes and Chromatosomes Visualized by Excessive-Pace Atomic Power Microscopy” by Bibiana Onoa, César Díaz-Celis, Cristhian Cañari-Chumpitaz, Antony Lee and Carlos Bustamante is licensed beneath a Inventive Commons Attribution 4.0 Worldwide License, which allows use, sharing, adaptation, distribution and copy in any medium or format, so long as you give applicable credit score to the unique creator(s) and the supply, present a hyperlink to the Inventive Commons license, and point out if modifications have been made. The photographs or different third occasion materials on this article are included within the article’s Inventive Commons license, except indicated in any other case in a credit score line to the fabric. If materials will not be included within the article’s Inventive Commons license and your meant use will not be permitted by statutory regulation or exceeds the permitted use, you will have to acquire permission instantly from the copyright holder. To view a replica of this license, go to http://creativecommons.org/licenses/by/4.0/.

